This project concerns the development of theoretical and computational methods to investigate rare structural transitions occurring in complex systems. To this end, we interface elements of non-equilibrium statistical mechanics, theory of stochastic processes, statistical field theory and renormalization group techniques with high-performance computing (HPC).
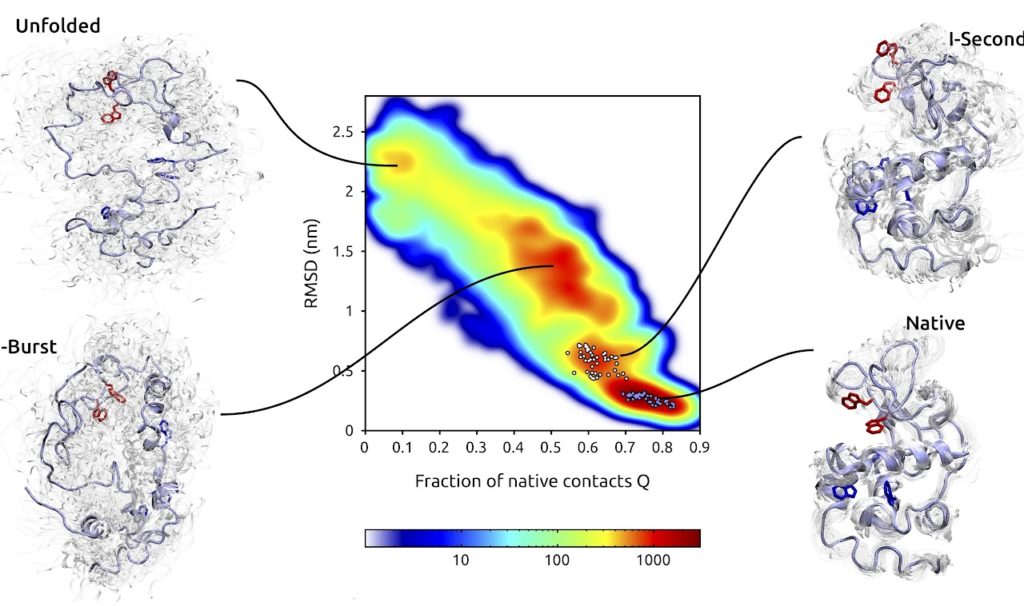
The most fundamental goal of this research is to understand the general physico-chemical mechanisms which drive rare structural transitions, in different conditions. In this context, our emphasis is on large conformational reactions undergone by biopolymers, in particular proteins. For example, in the paper of Ianeselli et al. in the list below we interface enhanced path sampling methods with quantum chemical electronic structure calculations in order to develop a scheme to interpret the results of time-resolved optical experiments.
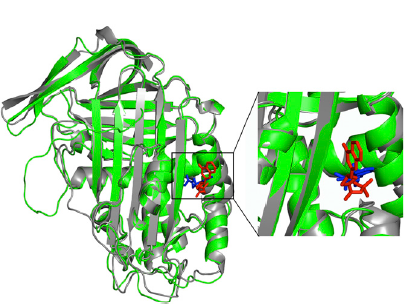
A second, more translational goal of our research is to apply the same theoretical and computational methods to model specific biological processes. For example, in the paper by Cazzolli et al. of the list below we use these techniques to clarify a conformational transition of a serpin protein which leads to its biological latency. Through these simulations we could explain why a specific small molecule is able to make this transition more frequent, contrasting pathologies associated to the over-expression of this protein in the cell.
REPRESENTATIVE BIBLIOGRAPHY
- Alan Ianeselli, Simone Orioli, Giovanni Spagnolli, Pietro Faccioli, Lorenzo Cupellini, Sandro Jurinovich, Benedetta Mennucci. The Atomic Detail of Protein Folding Revealed by an Ab-Initio Reappraisal of Circular Dichroism. J. Am. Chem. Soc. 140, 3674−3682 (2018)
- Simone Orioli and Pietro Faccioli. Dimensional Reduction of Markov State Models from Renormalization Group Theory., J. Chem. Phys. 145 124120 (2016)
- Simone Orioli , Silvio a Beccara , and Pietro Faccioli, Self-Consistent Calculation of Protein Folding Pathways. J. Chem. Phys. 147, 064108 (2017)
- Silvio a Beccara, Lorenzo Fant and Pietro Faccioli. Variational Scheme to Compute Protein Reaction Pathways Using Atomistic Force Fields with Explicit Solvent, Phys. Rev. Lett. 114, 098103 (2015)
- Giorgia Cazzolli, Fang Wang; Silvio a Beccara; Anne Gershenson; Pietro Faccioli, Patrick L. Wintrode. Serpin Latency Transition at Atomic Resolution Proc. Natl. Acad. Sci. USA 111, 15414 (2014).
- Silvio a Beccara, Tatjana Skrbic, Roberto Covino, and Pietro Faccioli. Dominant Folding Pathways of a WW Domain. Proc. Natl. Acad. Sci. USA 109, 2330 (2012)

