Single-Molecule Structure and Topology of Kinetoplast DNA Networks
Pinyao He, Allard J. Katan, Luca Tubiana, Cees Dekker, and Davide Michieletto Phys. Rev. X 13, 021010 – Published 19 April 2023
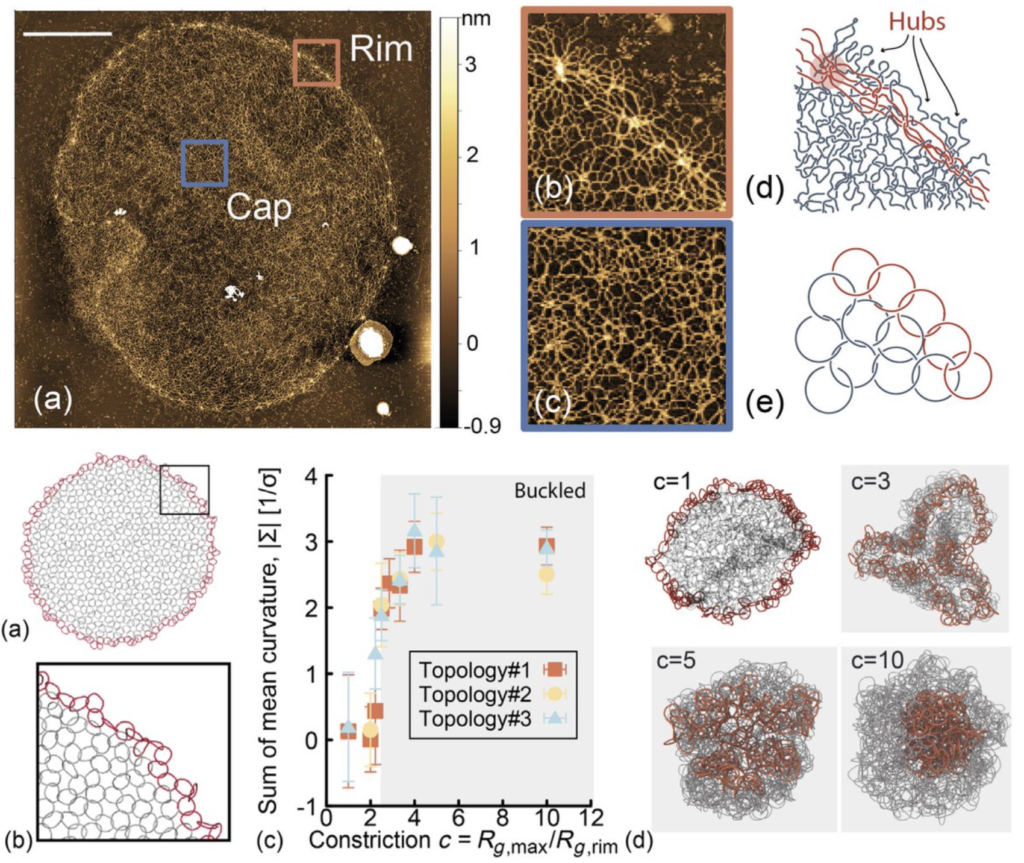
Trypanosomes are unique single-cell parasites: Their mitochondrial genome is made of thousands of DNA minicircles interlinked to form a giant 2D Olympic-ring-like network, known as a “kinetoplast DNA” (kDNA). This type of DNA organization is seen nowhere else in nature, and several questions on its evolution, self-assembly, and replication are far from answered. In this work, we combine high-resolution atomic force microscopy, quantitative image analysis, and molecular dynamics simulations to characterize the structure and topology of kDNAs at single-molecule resolution.
We show that the minicircles’ valence (the number of DNA rings linked to any one ring) has a broad distribution, with a mean around 3. We argue that the valence is controlled in vivo to drive the formation of a connected structure that preserves the integrity of the genome during replication yet avoids redundant constraints and a “topologically frustrated” rigid network. Our simulations explain that kDNAs undergo a buckling transition when extracted from the parasite, akin to the buckling of thermal elastic sheets. Finally, we estimate the bending rigidity and Young’s moduli of kDNAs to be thousands of times smaller than lipid vesicles. This ultrasoft nature of kDNAs may motivate the design of synthetic 2D interlocking ring networks, perhaps made of synthetic polycatenanes or DNA plasmids.
Our work sheds light on the structure and topology of a fascinating and topologically complex self-assembled genome at the single molecule scale and could inspire the creation of ultrasoft topological materials.

