M. Mele, R. Fiorentini, T. Tarenzi, G. Mattiotti, R. Potestio, Determining the optimal structural resolution of proteins through an information-theoretic analysis of their conformational ensemble
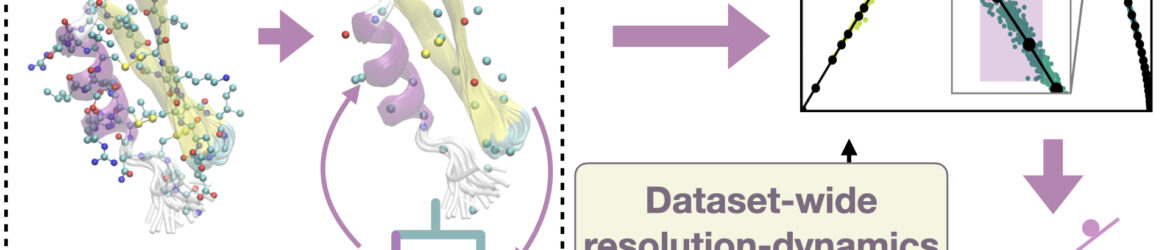
The choice of structural resolution is a fundamental aspect of protein modelling, determining the balance between descriptive power and interpretability. Although atomistic simulations provide maximal detail, much of this information is redundant to understand the relevant large-scale motions and conformational states. Here, we introduce an unsupervised, information-theoretic framework that determines the minimal number of atoms required to retain a maximally informative description of the configurational space sampled by a protein. This framework quantifies the informativeness of coarse-grained representations obtained by systematically decimating atomic degrees of freedom and evaluating the resulting clustering of sampled conformations. Application to molecular dynamics trajectories of dynamically diverse proteins shows that the optimal number of retained atoms scales linearly with system size, averaging about four heavy atoms per residue–remarkably consistent with the resolution of well-established coarse-grained models, such as MARTINI and SIRAH. Furthermore, the analysis shows that the optimal retained atoms number depends not only on molecular size but also on the extent of conformational exploration, decreasing for systems dominated by collective motions. The proposed method establishes a general criterion to identify the minimal structural detail that preserves the essential configurational information, thereby offering a new viewpoint on the structure-dynamics-function relationship in proteins and guiding the construction of parsimonious yet informative multiscale models.